GltS regulates biofilm formation in methicillin-resistant Staphylococcus aureus | Communications Biology - Nature.com
Abstract
Biofilm-based infection is a major healthcare burden. Methicillin-resistant Staphylococcus aureus (MRSA) is one of major organisms responsible for biofilm infection. Although biofilm is induced by a number of environmental signals, the molecule responsible for environmental sensing is not well delineated. Here we examined the role of ion transporters in biofilm formation and found that the sodium-glutamate transporter gltS played an important role in biofilm formation in MRSA. This was shown by gltS transposon mutant as well as its complementation. The lack of exogenous glutamate also enhanced biofilm formation in JE2 strain. The deficiency of exogenous glutamate intake accelerated endogenous glutamate/glutamine production, which led to the activation of the urea cycle. We also showed that urea cycle activation was critical for biofilm formation. In conclusion, we showed that gltS was a critical regulator of biofilm formation by controlling the intake of exogenous glutamate. An intervention to target glutamate intake may be a potential useful approach against biofilm.
Introduction
Biofilms are aggregates of bacteria that are adherent to a material surface and encased in a self-synthetic extracellular polymer matrix. Biofilm-related infections occupy a large percentage of nosocomial infections1, including device and indwelling catheter infections. Biofilm infections are highly resistant to antibiotic therapy and host defense, making biofilms difficult to be eradicated2. The transition of bacterial cells from planktonic to biofilm form is induced by the regulation of gene expression in response to sensed physicochemical environmental signals such as ions, pH, and nutrients, which in turn affects various signaling pathways3. Methicillin-resistant Staphylococcus aureus (MRSA) is a leading nosocomial pathogen and commonly associated with significant morbidity, hospital mortality, length of stay, and economic burden, with an estimated attributable healthcare cost of $1.7 billion in 2017 in the United States4. An investigation into the underlying molecular mechanisms of biofilm formation in MRSA is critical to developing a therapeutic approach.
The tricarboxylic acid (TCA) cycle is a central metabolic pathway that provides energy, reducing potential, and biosynthetic intermediates. During biofilm formation, the TCA cycle may be initially activated5, but the inactivation of the TCA cycle generally ensues6, contributing to an increase in the percentage of protein components in the USA300 biofilm matrix7,8. The involvement of the urea cycle in biofilm formation is also described7. Ion transporters play a critical role in sensing environments and regulating ion trafficking. We have previously shown that the a subset of ion transporters was involved in biofilm formation in Escherichia coli9. The biological roles of ion transporters in MRSA were recently studied by several investigators10,11,12,13. For example, alsT functions as an efficient glutamine transporter13 and gltT is an aspartate transporter11. However, their role in biofilm has not been extensively studied. With the hypothesis that a subset of MRSA ion transporters would be involved in biofilm formation, we screened biofilm formation for ion transposon (Tn) mutants in the Nebraska library14. We found significantly increased biofilm formation in a major sodium-glutamate symporter gltS transposon mutant strain (gltS::Tn)13 compared to the parental strain. Because the gltS mutant was the only one that demonstrated alteration in biofilm formation among all the sodium ion transporters, we tested our hypothesis that glutamate-induced metabolic changes would affect MRSA biofilm formation by utilizing chemically defined media (CDM) depleted of glutamate, mass spectrometry-based metabolomic analyses, and RNA transcriptomic analyses. Although the involvement of the TCA and urea cycles in biofilm formation has been previously described as above7,15,16, it is not known how exogenous glutamate, a major amino acid in central metabolism, is involved in biofilm formation in conjunction with the TCA and urea cycles. Understanding the role of glutamate uptake via gltS in MRSA biofilm formation will provide an insight into mechanisms that regulate biofilm formation.
Results
GltS transposon mutant enhanced biofilm formation in vitro
To test the hypothesis that a subset of ion transporters would be responsible for biofilm formation, we first performed screening experiments using a 96-well format crystal violet-based biofilm assay. The list of Nebraska library strains that we tested is shown in Supplementary Data 1, and the results are shown in Fig. 1a. Among the 20 transposon mutants tested, gltS transposon mutant (gltS::Tn) significantly enhanced biofilm formation compared to the parent strain JE2. Potassium-transporting ATPase ATP-binding protein mutant (kdpB::Tn) also showed enhanced biofilm formation compared to that of the parent strain, although less than gltS::Tn (Fig. 1a). The growth was similar for the parent and gltS::Tn strains (Fig. 1b), indicating that bacterial growth rates did not account for the phenotype. We also examined the growth of all the other mutants we tested in Fig. 1a. No mutants showed any significant difference in growth compared to their parent strain (Supplementary Data 2). We examined biofilm mass by assessing live/dead cell-stained samples in two-well plastic chamber slides. The z-stack images of the biofilm at 2.45 μm intervals showed cells deposited in the biofilm at a depth of approximately 22.05 μm for the parental strain and 36.75 μm for the gltS::Tn strain. (Fig. 1c). The volume of biofilm calculated by using the voxel counter per same surface area for gltS::Tn was significantly larger than the one of JE2 (Fig. 1c), consistent with the results of the crystal violet-based assay (Fig. 1a). The thickness data also corresponded to the volume (Fig. 1c). The thickness of live cells in gltS::Tn was significantly larger than in JE2. To confirm that the increased biofilm formation of the gltS::Tn mutant was due to the gltS deficiency and not due to any secondary mutations in the genome, the complementation of gltS was performed. The biofilm formation of the complemented strain was comparable to that of JE2, indicating that gltS deficiency was responsible for biofilm enhancement (Fig. 1d).
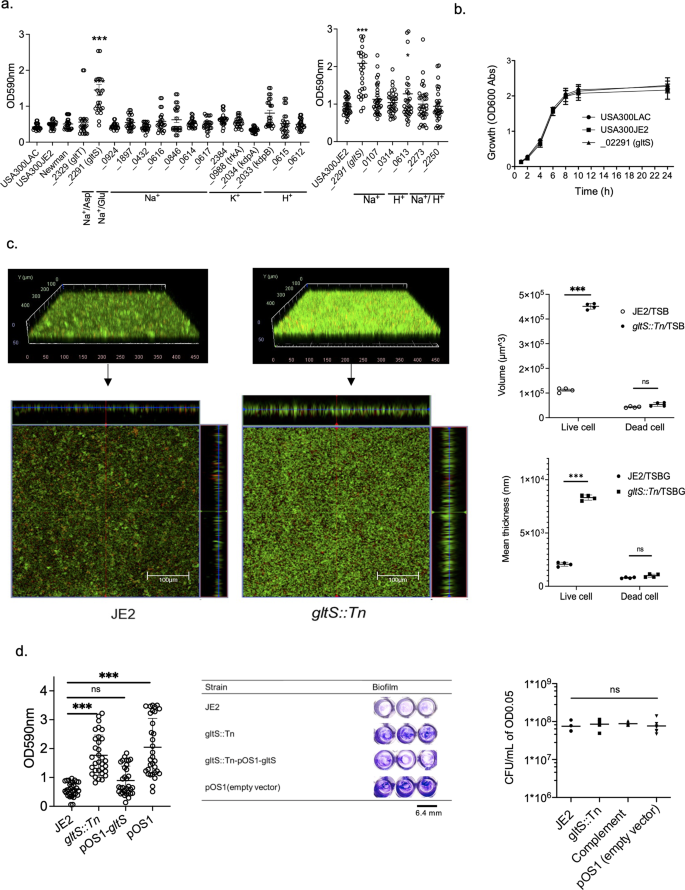
a Crystal-violet based biofilm screening of ion transporter mutants. Biofilm quantification in 1% glucose TSB medium at 48 h. We included accession number for all the genes. For one that have specific gene name, we included it in parenthesis. For example, gltT has gene accession number of SAUSA300_2329. In this case, we show as _2329 (gltT). If we do not have gene number such as gene with accession number of SAUSA300_0924, we just show accession number as _0924. Data are shown as mean ± SD of three independent tests with 8 replicates. One-way ANOVA with post hoc Bonferroni's multiple comparisons test vs USA300 JE2. ***p < 0.001, *p < 0.05. b Growth curves of USA300 parental strain, and gltS mutant strain by OD600 absorption value of turbidity. c (Left) Orthogonal, z-stacked images of biofilms with a confocal laser microscope. The 48-h biofilm was subjected to live/dead staining. Live cells are shown in green (488 nm excitation laser) and dead cells are shown in red (570 nm wavelength filter). A representative image of two independent experiments is shown. The volume of biofilm calculated by using the voxel counter plugin in Image J and shown as mean ± SD of four independent locations. One-way ANOVA with post hoc Bonferroni's multiple comparisons test vs USA300 JE2. ***p < 0.001. d Crystal violet-based biofilm assay using complemented ΔgltS strain (gltS::Tn-pOS1-gltS) at 48 h. One-way ANOVA with post hoc Bonferroni analysis. ***p < 0.001. (Middle) OD measurement of eight replicates is shown as mean ± SD. Crystal violet-stained Images. (Right) Because we used solution with O.D. of 0.05 as an input for biofilm assay, we compared CFU with O.D. of 0.05 for each strain. Data is shown as mean ± SD of 6 replicates. Statistical significance was tested using one-way ANOVA with post hoc Bonferroni analysis. No difference was observed.
To evaluate the differences in biofilm formation and components among the strains, we compared the expression levels of genes involved in biofilm component and performed biofilm assays using each reagent treatment on biofilm supernatants. The biofilms of the parental strain and gltS::Tn were reduced to a similar degree by proteinase K treatment (Supplementary Fig. 1a, 56.8%, and 66.8% reduction, respectively). There was a significant reduction in the amount of gltS::Tn biofilm after DNase treatment at 24 h, not in the parent strain (Supplementary Fig. 1b). At 48 h, DNase treatment attenuated gltS::Tn biofilm amount, although not statistically significant. Not all eDNA could be digested by the DNase I test, since several types of DNA-binding proteins in S. aureus, such as phenol soluble modulin (PSM) peptides and Eap, can protect eDNA from digestion by DNase I17. Although we did not find any difference in the percentage of protein among biofilm constituents, the type of proteins is important in biofilm formation.
GltS::Tn mutant enhanced biofilm formation in vivo
To examine the role of gltS in vivo, we sought for an in vivo biofilm formation model. Deep muscle infection model is a model created by inoculating MRSA, followed by muscle suture closure as well as skin closure (Fig. 2a). In both JE2 and gltS::Tn strains, the biofilm formation was seen on the muscle sutures by electron microscopy18 (Fig. 2b). Histology analysis demonstrated more extensive leukocyte infiltration following gltS::Tn infection (Fig. 2c). Bacterial loads of both muscle suture and muscles showed that gltS::Tn infection was associated with a significantly greater bacterial burden (Fig. 2d), in line with our in vitro data. IL-6 is one of the indicators for organ injury. IL-6 level was higher in gltS::Tn treated wound (Fig. 2e), suggesting more tissue damage from gltS::Tn infection. Lastly we examined glucose level in the wound. Glucose level at the wound site infected with JE2 or gltS::Tn showed a tendency to be lower than the baseline, but the difference was not statistically significant (Fig. 2f).
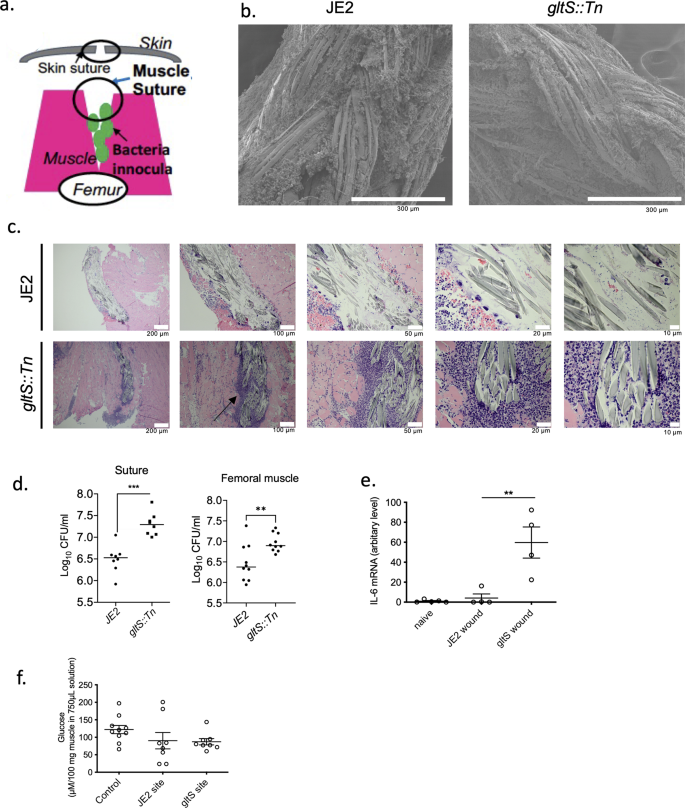
a Scheme of a murine wound infection model. b SEM image of muscle sutures. Representative images from two independent experiments. Scale bar (white) in each image. c Hematoxylin and eosin (H&E) staining of the surgical infection site. Scale bar (white) in each image. A black arrow indicates extensive leukocyte infiltration. d Sutures were sonicated in the same volume of PBS for each strain, and the CFUs were counted. Muscle sections of the same weight from each mouse were homogenized, and the CFUs in the homogenized suspensions were quantified. Data are shown as mean ± SD of 8 mice for suture and 10 mice for muscle sample. e IL-6 mRNA levels in the wound tissues at 48 h after infection with JE2 or gltS::Tn were examined using RT-qPCR. Data are shown as mean ± SD of 4 replicates. One way ANOVA with post hoc Bonferroni analysis was done. **p < 0.01. f Glucose levels at the wound were measured. Data are shown as mean ± SD of 6 replicates. One way ANOVA with post hoc Bonferroni analysis was done. No statistical significance was observed.
Exogeneous glutamate deficiency enhanced biofilm formation
GltS is a symporter of Na/glutamate from the environment to the intracellular space. Because gltS::Tn mutant was the only one that showed increased biofilm formation among all the Na transporters tested, we hypothesized that exogenous glutamate intake plays a significant role in biofilm formation. A previous study showed that gltS::Tn impaired glutamate intake, while gltT::Tn (strain 2329) did not show any reduction in glutamate transport13. Accordingly, we examined the role of exogenous glutamate in biofilm formation of JE2 parent strain using a chemically defined medium + 1% glucose (CDM) and CDM depleted of glutamate. Glutamate depletion led to a significant increase in biofilm formation assessed by both the crystal violet method (Fig. 3a) and confocal microscopy (Fig. 3b), in line with the gltS::Tn phenotype. We measured the number of viable biofilm and planktonic cells using live/death staining using fluorescence intensity as a surrogate. There was no significant difference in cell number between complete CDM and glutamate-depleted medium (Fig. 3c). The results suggested that exogenous glutamate was not essential for cell viability under 48-h culture conditions.
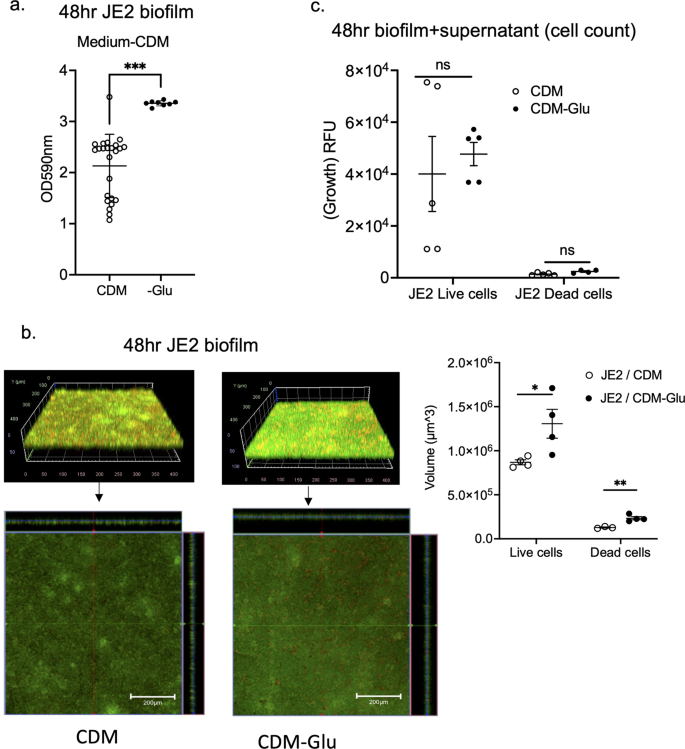
a Absorbance values of crystal violet assays of 48 h JE2 biofilms using complete CDM containing glucose medium (CDM) and glutamate (Glu)-depleted from CDM medium (CDM-Glu). Data are shown as mean ± SD of 8 replicates. Student t test was performed. ***p < 0.001. b Images of JE2 biofilm after LIVE/DEAD staining by CLSM and channel-specific volume calculated by Image J. The volume of biofilm calculated by using the voxel counter plugin in Image J and shown as mean ± SD of four independent locations. One-way ANOVA with post hoc Bonferroni's multiple comparisons test vs USA300 JE2. * and **p < 0.05 and p < 0.01. c Cell counts were determined by using fluorescence intensity of the whole 48-h biofilm and supernatant as a surrogate. Fluorescence intensity was measured as relative fluorescence unit (RFU) because the spectra of collected light should be corrected for instrument effects. Data are shown as mean ± SD of 8 replicates. One-way ANOVA with post hoc Bonferroni's multiple comparisons test vs USA300 JE2. No statistical significance was observed.
Intermediate metabolites of the urea cycle and TCA cycle were more abundant in the gltS::Tn mutant
Because our data so far showed an inverse correlation between glutamate intake and biofilm formation, we examined the metabolic derangement induced by gltS::Tn using a metabolomics analysis. All data in Fig. 4 were from metabolite analysis in biofilm and supernatant using TSB as medium. Figure 4a shows the principal component (PC) analysis; the total contribution of PC1 and PC2 was relatively high, 77.4% in bacterial cells and 71.9% in supernatant. For cells, mevalonic acid, glucose-1-phosphate, and lysine were major contributors to PC1, while succinic acid, β-alanine, and ornithine were major contributors for PC2. On PCA analysis, the supernatant profiles of JE2 and gltS::Tn biofilm conditions almost overlapped with each other. Planktonic and biofilm cells showed a clear difference, while a separation of the intracellular metabolite profiles between JE2 and gltS::Tn biofilm was limited.
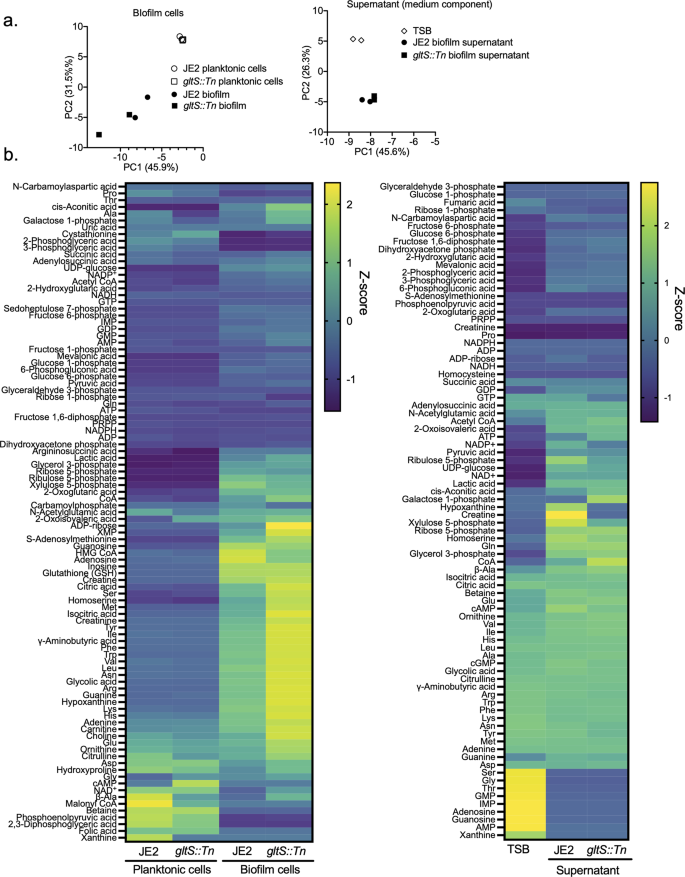
a Principal component analysis (PCA) of metabolites in biofilm cells (left) and supernatant, showing the top two on the x-axis and y-axis, respectively. For supernatant analysis, TSB indicates TSB containing 1% glucose without any bacteria co-culture. b Heatmap representation of metabolome profiles analyzed by hierarchical clustering.
The results of all the measured metabolite profiles were shown by a heatmap (Fig. 4b). Details of pathways will be discussed later; Fig. 4b mainly showed that biofilms in TSB produced a majority of the amino acids, including glutamate, glutamine, and arginine, about 1.4- to 2.0-fold more in gltS::Tn than in the parent strain. The absolute concentrations of all metabolites measured are shown in Supplementary Data 3, which includes data from using CDM and glutamate-depleted CDM as medium.
Since parameters such as energy charge, lactate-to-pyruvate ratio, glutathione redox ratio, and total amino acids are useful for gaining further insights into physiological states such as energy and redox status, we listed equations to calculate each metabolic parameter, its relevance to cell metabolism and physiological states, and results in Supplementary Data 4. NAD+/NADH ratio indicates metabolic activity19. The NAD+/NADH ratio of gltS::Tn/JE2 strains was 1.6, indicating higher metabolic activity in gltS::Tn. This is because NAD+, a cofactor in many reactions such as glycolysis, the TCA cycle, and beta-oxidation of fatty acids, was 1.6 times more abundant in gltS::Tn biofilm cells than in the parental strain (Supplementary Data 3). The glycerol 3-phosphate/dihydroxyacetone phosphate (DHAP) ratio serves as an indirect parameter for an inverse ratio of NAD+/NADH and energy status. The relative ratio of gltS::Tn/JE2 strains was low (0.5).
The analyzed metabolites were subjected to metabolic pathway analysis using MetaboAnalyst 5.0 online (Supplementary Data 5). Metabolites with an absolute difference of more than 0.5 in the relative peak area used for metabolome profiling between planktonic cells and 48-h biofilms were included in the analysis. This led to the creation of a heatmap showing metabolites grouped by pathway, focusing on purine metabolism, arginine biosynthesis, pentose phosphate (PP) pathway, and citrate cycle besides the glutamate metabolism (Supplementary Fig. 2a, b). Metabolites generated in the TCA cycle were overall produced in greater abundance in the biofilm state with the rank order of gltS::Tn > JE2 strain (Supplementary Fig. 2a). Regarding 2-oxoglutarate (2-OG), there was almost no difference between strains, while glutamate/2-OG ratio was 1.3 times more in gltS::Tn compared to JE2. Coenzyme A (CoA) was seen more in gltS::Tn cells and supernatant. Similarly, urea cycle related metabolites such as ornithine, citrulline, argininosuccinic acid and arginine were higher in gltS::Tn biofilm cells (Supplementary Fig. 2b). Creatine was higher in the supernatant of JE2 strain compared to gltS::Tn. As for the PP pathway/glycolysis, the amount of metabolites produced from the planktonic cells relative to the biofilm was higher under PP pathway than under glycolysis (Supplementary Fig. 3a). AMP, hypoxanthine, adenine, XMP, and guanine in purine pathway were produced in higher amounts in gltS::Tn (Supplementary Fig. 3b, Supplementary Data 5), in line with higher levels of glutamate and glutamine in the mutant.
The expression of urea cycle-mediated genes was significantly upregulated in gltS::Tn
Based on the metabolites data described above, we focused on analyzing the transcriptomic profiles of genes involving in the TCA and urea cycles. The genes of enzymes involved in the TCA cycle (acnA, icd, and sucC) were universally downregulated in biofilm cells compared to planktonic cells in both strains (Fig. 5). Although the levels of TCA cycle-mediated metabolites were higher in biofilm cells than in planktonic cells (Supplementary Fig. 2a), RT-qPCR result indicated the presence of inhibitory mechanism for TCA cycle activity. Although TCA cycle-related metabolite levels were higher in gltS::Tn mutant compared to the parent strain, there was no significant difference in mRNA expression of the tested genes between the strains. We performed mass spectrometry analysis of JE2 and gltS::Tn planktonic and biofilm cells, and compared their TCA cycle related enzyme levels. Some of the TCA cycle related enzymes in gltS::Tn cells had higher levels at 24 h. The enzyme levels in gltS::Tn cells were consistently higher than those in JE2 strain at 48 h (Supplementary Data 6), consistent with the metabolomics data (Fig. 4). gltB and gltD, which encode the subunits of glutamine oxoglutarate aminotransferase (GOGAT) and are involved in glutamate and glutamine metabolism, were expressed significantly higher in gltS::Tn than in the parental strain at 0 and/or 4 h (Fig. 5), which explained higher glutamate and glutamine levels in gltS::Tn biofilm cells. This enzyme was not consistently detected in our mass spectrometry data set.
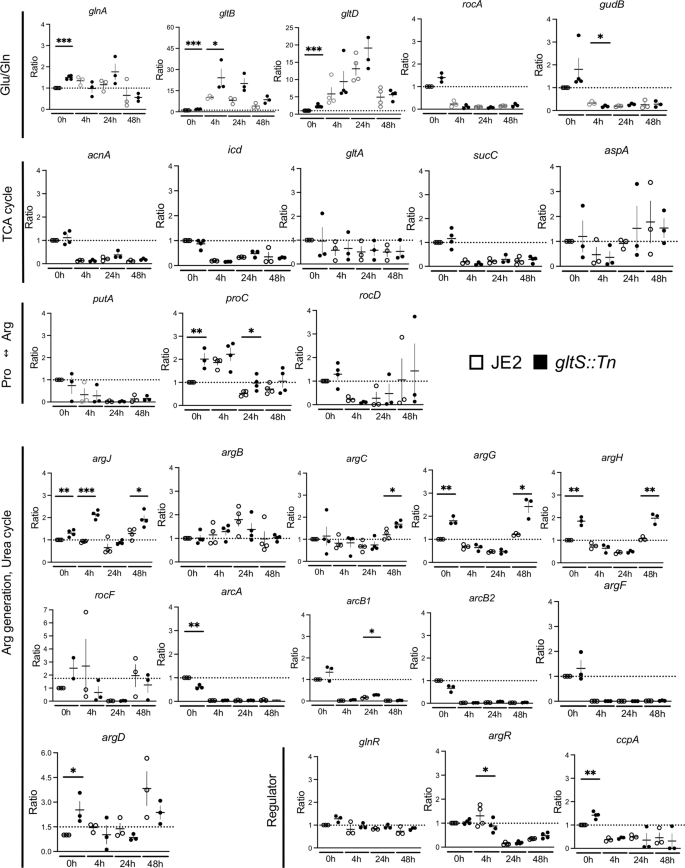
Expression ratio based on the 0-h value of the JE2 strain over three independent tests. 0 h corresponds to planktonic cells, while 4 h, 24 h and 48 h correspond to biofilm cells at 4, 24, and 48 h-incubation, respectively. Data are mean ± SD of 3–4 replicates. Student t-test between strains of samples collected at the same time. *P < 0.05, **P < 0.01, ***P < 0.001.
Glutamate can be shuttled and available for urea cycle metabolism. Our data also showed gene expression of arginine producing molecules were overall upregulated in gltS::Tn. An upregulated expression of argJ (bifunctional glutamate N-acetyltransferase/amino-acid acetyltransferase) and argCD in gltS::Tn suggests that glutamate to arginine influx significantly increased, consistent with the result of the metabolic pathway data (Supplementary Fig. 2b). The expression of argR, an arginine repressor, was significantly lower in gltS::Tn than in the parental strain in 4 h-biofilm cells. These data further demonstrated more active use of the urea cycle in gltS::Tn cells.
RNA sequencing analysis for additional pathway consideration
We applied RNA sequencing experiments (RNA seq) to analyze the expression of genes in pathways other than those examined by RT-qPCR, as well as to understand the expression of supplementary gene subsets associated with the pathways of the focus (Supplementary Fig. 4). Genes that were at least 2-fold more (or less) expressed by RNAseq analysis in gltS::Tn strain relative to the parental strain at the time of each cell harvest are shown in Supplementary Data 7.
First, we examined TCA cycle and urea cycle related genes. The heatmap shows the log2 fold change of each gene at indicated time points compared to time point 0 h in each pathway (Fig. 6). TCA cycle-related genes were downregulated in biofilm cells, while arginine production-related genes were upregulated, consistent with our RT-qPCR data. The expression of arginine deiminase20, which converts arginine to citrulline and is highly expressed during biofilm growth in multiple bacterial species21, peaked at 24 h in both strains, and was higher over time in gltS::Tn. Urease is also associated with biofilm formation22. The expression of ureAB in gltS::Tn (0 h) was more than twice that of the parental strain (Supplementary Data 7).
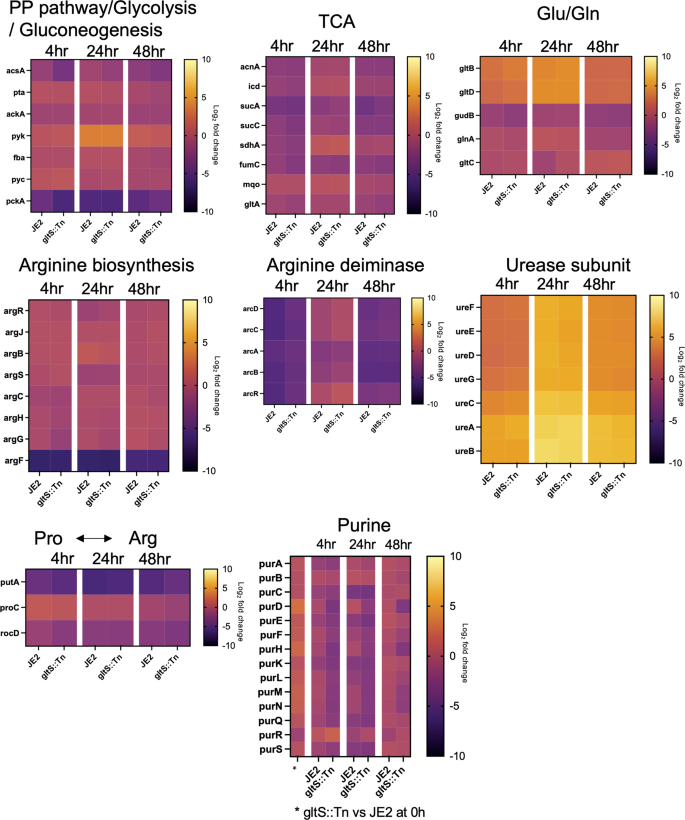
Heat map showing the course of change in log2 fold change at each time point referenced to the start of biofilm culture (0 h). The bar indicates log2 fold change at each time point referenced to the start of biofilm culture (0 h).
Glutamate production pathway-related genes were also upregulated in the mutant strain. gltC positively regulates the expression of GOGAT in Bacillus subtilis. Its MRSA homolog in the gltS mutant was expressed ~2-fold higher than JE2 at 24 h. Among genes responsible for the pathway that generates glutamate from histidine, hutI and hutU were found to be upregulated in gltS::Tn at time 0 (Supplementary Data 7), which might indicate extra glutamate production pathways in addition to GOGAT.
Interestingly, the expression of ahpFC, which serves to protect cells from DNA damage by alkyl hydroperoxides, was higher in the mutant strain at all the four time points (Supplementary Data 7). In addition, at the beginning of biofilm culture or at an earlier time point, presumably before attachment, genes related to reactive oxygen species (ROS), heat shock regulation, biofilm cell attachment, and purine production, such as dps, grpE, clpC, sdrD and purHLMN, were expressed at higher levels in gltS::Tn. These data are consistent with the higher biofilm formation of the gltS::Tn strain.
Lastly, the expression of the purine production pathway was higher in gltS::Tn only at time point 0 h, and the trend was reversed after 4 h (Fig. 6). Interpretation of this result needs further study.
Determination of cyclic-di-AMP levels and dacA and gdpP gene expression
Cyclic-di-AMP (c-di-AMP) is an intracellular signaling molecule involved in various functions including ROS production, biofilm formation, and negative regulation of the TCA cycle, particularly in Gram positive bacteria13,23,24. Intracellular c-di-AMP levels of the parental strain significantly increased during biofilm formation. At 48 h, it exceeded by 10-fold that of JE2 planktonic cells (Fig. 7a), which correlated with the previous reports describing an association between c-di-AMP and biofilm formation23,25. c-di-AMP negatively regulates the TCA cycle, and our gene expression analysis of JE2 indicated the negative regulation of TCA cycle during biofilm formation (Fig. 5). Intracellular c-di-AMP levels of gltS::Tn showed a similar trend. However, at 24 h, c-di-AMP level of gltS::Tn mutant was reduced by ~50% compared to that of the parental strain. Because glutamine is one of contributors to reduce c-di-AMP levels13, higher glutamine in gltS::Tn: matches with this c-di-AMP result. This lower c-di-AMP level in gltS::Tn is also in line with higher TCA metabolite production in gltS::Tn. Interestingly, by 48 h, the c-di-AMP levels in the two strains were comparable. The expression of a c-di-AMP-degrading phosphodiesterase gdpP in gltS::Tn was approximately twice as high as that of the parental strain (Fig. 7b). dacA expression did not show a significant difference between the two strains (Fig. 7b). gdpP expression level was higher in gltS:Tn, consistent with lower c-di-AMP levels in gltS mutant (Fig. 7b).
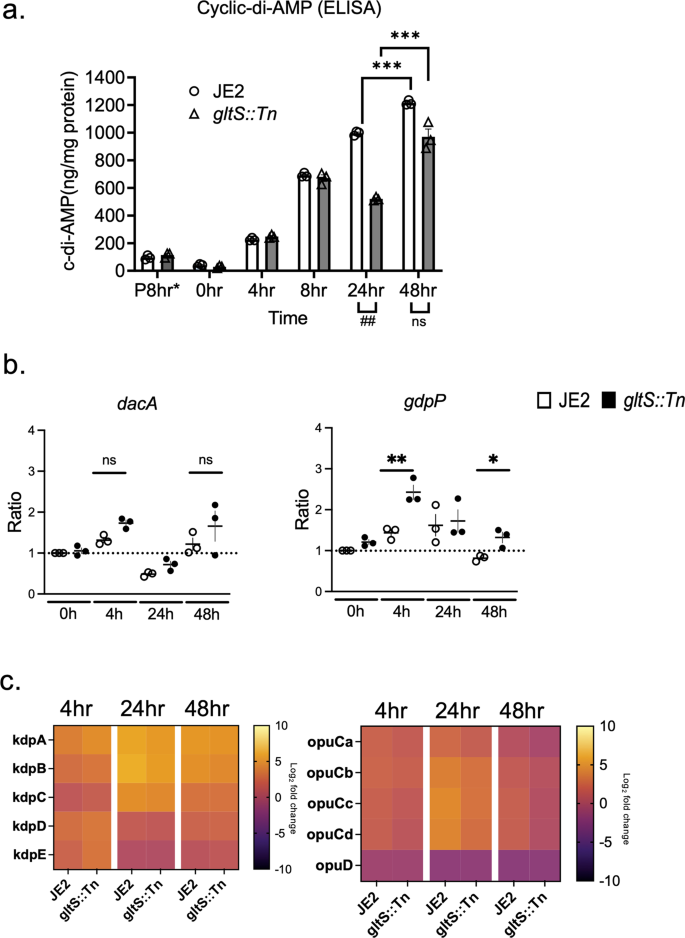
a Intracellular cyclic-di-AMP levels measured by ELISA. Data are shown as mean ± SD of 3 replicates. Student t-test was used. ***p < 0.001. *P 8 h indicates 8 h after starting planktonic culture. b Gene expression ratio of dacA and gdpP by RT-qPCR. Data are mean ± SD of 4 replicates. Student t-test between strains of samples harvested at the same time. *P < 0.05, **p < 0.01. c Heatmap of RNA seq results for kdp and opu operons. Log2 fold change from expression at time 0 for each gene. The bar indicates log2 fold change at each time point referenced to the start of biofilm culture (0 h).
Glutamate is a major intracellular counter ion for potassium26. c-di-AMP binds to K exporters and plays a significant role in osmolarity regulation, mainly by modulating the uptake of potassium and osmolytes27. We used RNA seq data of genes involved in potassium channel and osmolytes uptake (kdp operon and opu operon, respectively) to show the time course of their expression ratios from time 0 in a heatmap (Fig. 7c). Overall, kdpA-E expression was upregulated during biofilm formation in both strains. In our screening, kdpB::Tn demonstrated enhanced biofilm formation (Fig. 1a). The expression of opuCa and opuD was downregulated particularly at 48 h when the c-di-AMP level was highest, with the expression of gltS::Tn being lower.
A study using mutants related to arginine synthesis and the urea cycle: the role of citrulline
With the hypothesis that accelerated urea cycle metabolism would be responsible for enhanced biofilm formation in gltS::Tn, we performed a biofilm assay with Tn mutants related to arginine/urea metabolism (Fig. 8a). Although not statistically significant, argF::Tn markedly reduced biofilm formation, while argG::Tn, argJ::Tn, and arcB::Tn showed significantly enhanced biofilms. No growth difference among the strains was observed (Supplementary Data 2). Because argF is an enzyme that mediates the synthesis of citrulline from ornithine, and argG and arcB mutants enhanced the biofilm, we hypothesized that citrulline would be essential for the biofilm and added citrulline to the TSB culture medium. Both the parental strain and argF::Tn showed a significant increase in biofilm formation in a citrulline concentration-dependent manner, although the amount of biofilm formation was not as high as that of gltS::Tn (Fig. 8b). Since TSB contains about 50 μM citrulline, we also performed the assay using CDM with or without citrulline. These results were comparable to those of TSB. These results suggest that citrulline would be an important mediator for biofilm formation, and higher citrulline level in gltS::Tn (Supplementary Fig. 2b) might have been at least in part responsible for its biofilm phenotype. The mechanism of biofilm enhancement by argJ::Tn is unclear. Proline can be a source of citrulline. Thus, we examined the role of proline depletion in biofilm formation. Proline depletion attenuated biofilm formation (Fig. 8c). We also examined the biofilm formation using TCA cycle gene transposon mutants (Supplementary Fig. 5). They did not affect biofilm formation, indicating the involvement of urea cycle in biofilm. Tn mutant of glutamate synthase, gltD::Tn, also significantly increased the biofilm formation.
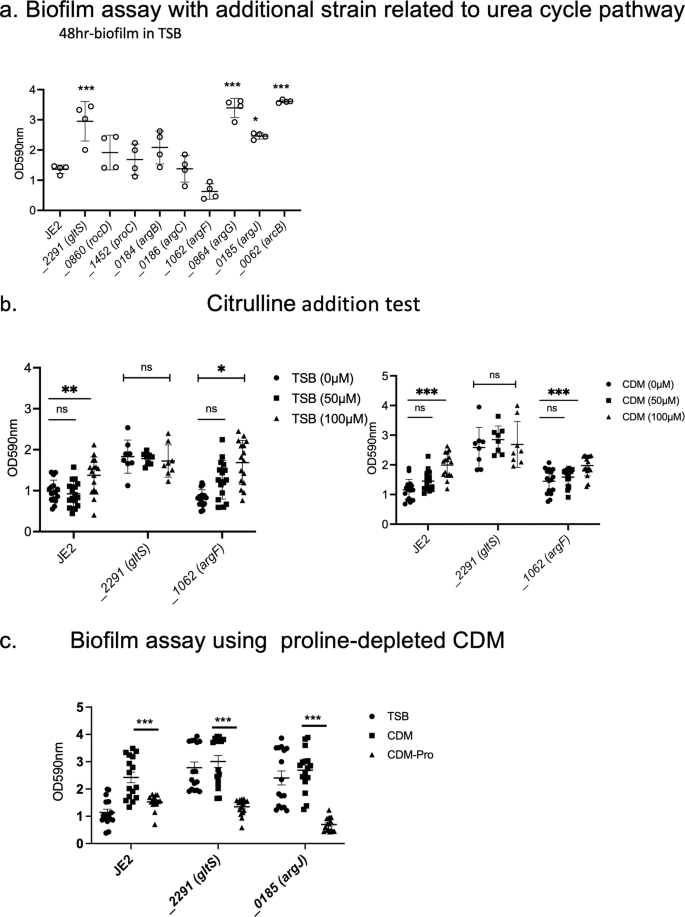
a Biofilm assay of urea cycle pathway related gene transposon mutants. Biofilm quantification in 1% glucose TSB medium at 48 h. Data are shown as mean ± SD of 8 replicates. One-way ANOVA with post hoc Bonferroni's multiple comparisons test vs USA300 JE2. ***p < 0.001, *p < 0.05. b 48-h biofilm assay using the medium in the presence of additional citrulline (50 µM, 100 µM). Data are shown as mean ± SD of 8 replicates. One-way ANOVA with Bonferroni post hoc analysis. *P < 0.05, **P < 0.01, ***P < 0.001. c Biofilm assay using proline-depleted CDM. OD value of 48-h biofilm. Data are shown as mean ± SD of 8 replicates. One-way ANOVA with Bonferroni post hoc analysis. ***P < 0.001.
Discussion
Staphylococcus aureus has evolved remarkably to be able to adapt to complex environmental changes in the host, and it forms colonies in a variety of niches28. One particularly important example of bacterial adaptation is the ability to grow as part of a sessile community of biofilm. A biofilm is a functional multilayered community of microorganisms, adhering to a surface and organized within a self-produced exopolymeric matrix29. Its complex regulation is based on drastic metabolic changes in bacterial cells. In this study, we found that MRSA biofilm formation was enhanced by inactivation of gltS. The summary of our findings is shown in Fig. 9.
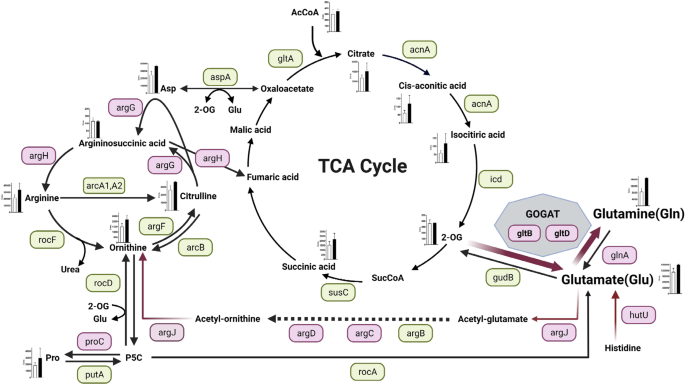
Red indicates that the expression of the gene was higher than that of the parent strain by RT- qPCR at any of the observation times from 0 to 48 h, and green indicates that there was no significant difference between the strains. However, even if there is a significant difference, the expression ratios of both strains that are less than 1-fold of the reference (JE2 time 0) are omitted and shown in green. For hutU, RNAseq data showed that expression was significantly (p < 0.05) higher than 2-fold in the gltS::Tn strain, hence it is additionally shown in pink. Absolute values of the concentration of each metabolite in the cells of 48-h biofilms are shown in the bar graph. White indicates the parent strain and black indicates gltS::Tn strain.
Glutamate and glutamine serve as the major amino acid donors for all nitrogen-containing compounds in all living organisms, including other amino acids and components of DNA and RNA synthesis. It has been suggested that 80–88% of the nitrogen incorporated into biomass is supplied by glutamate30. Therefore, the glutamate pool in cells needs to be kept high31. The intracellular glutamine/glutamate ratio serves as an important indicator of nitrogen availability in bacterial cells32. In S. aureus, there are several pathways to produce glutamate. In gltS::Tn, a rapid and intense activation of GOGAT was the primary response to impaired exogenous glutamate uptake for intracellular glutamate pool restoration. The concentrations of both glutamate and glutamine were higher in the gltS::Tn strain in the 48 h biofilm. The intracellular glutamate concentration is usually higher than the glutamine concentration because excess glutamine is converted to glutamate via the GOGAT pathway by glutamine oxoglutarate aminotransferase, which is composed of gltB and gltD protein subunits in S. aureus30. RNA seq data suggested that histidine to glutamate conversion pathway might also contributed to endogenous glutamate generation in gltS::Tn. Furthermore, asparagine serves as an important amino acid for glutamate biosynthesis as shown in Fig. 9, but aspA expression level did not differ between the strains, suggesting its limited role of this pathway in endogenous glutamate production by gltS::Tn.
The role of GOGAT was extensively studied in Bacillus subtilis, a model Gram-positive bacterium. GOGAT (gltAB) produces two molecules of glutamate from glutamine and 2-OG33. Expression of the gltAB gene is triggered by signals originating from carbon and nitrogen metabolism and is dependent on the LysR-type transcriptional activator gltC and the global regulator protein of nitrogen metabolism TnrA34. In the presence of glucose, gltC activates transcription of the gltAB operon to respond to an increased need for glutamate35. On the other hand, in the presence of glutamate, gltC is inactive and cannot activate the expression of the gltAB operon30. It is possible that gltC is activated when glutamate uptake is reduced. Although S. aureus gltC is present, its function is not known36. In a 24-h biofilm, we observed that gltC was expressed two-fold higher in gltS::Tn compared to the JE2 parent strain. However, the relationship between gltC and GOGAT in USA300 remains to be determined.
According to a nuclear magnetic resonance analysis, amino acids other than arginine, histidine, and proline must go through the TCA cycle and GOGAT to produce glutamate37. Our data showed increased TCA cycle metabolite levels. TCA cycle receives input at various steps. Metabolite profiles showed that the amount of PP pathway into the TCA cycle...

Comments
Post a Comment